Disclaimer: This article is more than two years old. Developments in science and computing happen quickly, and more up-to-date resources on this topic may be available.
The 2022 International Conference on Computational Science (ICCS) has awarded the Best Main Track Paper to “Establishing Metrics to Quantify Underlying Structure in Vascular Red Blood Cell Distributions,” co-authored by Duke University graduate student Sayan Roychowdhury, Center for Applied Scientific Computing (CASC) computer scientist Erik Draeger, and former Lawrence Fellow Amanda Randles, now a professor at Duke.
The paper offers a quantitative approach to systematically comparing computational blood flow models. Accurately simulating the dynamics of cell motion and interactions within the microvasculature is a problem of magnitude: How many spatial arrangements of cells will ensure meaningful results? The team initially thought they would find a solution by searching the scientific literature. “However, it turned out that no one seemed to have solved the specific problem [of packed objects flowing through a fixed length] we had,” states Draeger, who serves as leader for CASC’s Scientific Computing Group and director of LLNL’s High Performance Computing Innovation Center.
For Roychowdhury, who will complete a PhD in Biomedical Engineering in 2023, the award motivates him to continue his research at the intersection of biomedicine and high performance computing (HPC). He says, “I’m thankful that my advisor [Randles] was extremely supportive of this work because it started off as a small offshoot from another project. To see it come together and lead up to the Best Paper Award is very rewarding.”
Characterizing Ensemble Behavior
Crammed inside a human’s 60,000 miles of blood vessels are more than 25 trillion red blood cells (RBCs). The path of a tumor cell moving through this crowded space depends on the specific neighborhood of RBCs it encounters. “Imagine working your way through a crowded party without bumping into anyone, or stepping out of the way if you do make contact,” Draeger says. “The way you move through the room is very different depending on who else is there and where they’re standing.”
Therefore, predicting a tumor cell’s path—and where it is likely to reach a vessel wall to establish a secondary tumor site—is not as straightforward as simulating one arrangement of RBCs. Many cell configurations are needed to determine convergence of all possible behaviors, but how many configurations are enough? The team’s method enables researchers to identify well-generated, randomly placed cell distributions and to quantify distinct cell configurations. To do this, they used specific metrics to characterize a set of configurations that represents cells’ ensemble behavior.
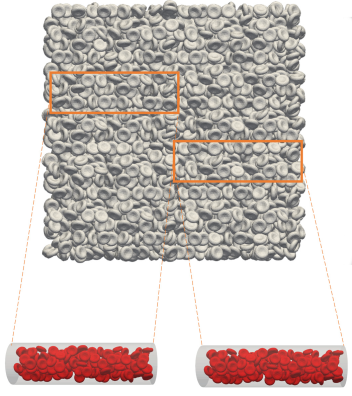
First, the team generated a large geometric system of packed cells and “submerged” a three-dimensional blood vessel in it, using the resulting filled vessel as the starting point for a simulation. (See Figure 1.) Individual configurations of thousands of cells were made by refilling the vessel at different locations in the bulk system. They then applied the radial distribution function—commonly used to calculate randomness in a dense distribution—to these arrangements to confirm the cells’ random placement and spacing.
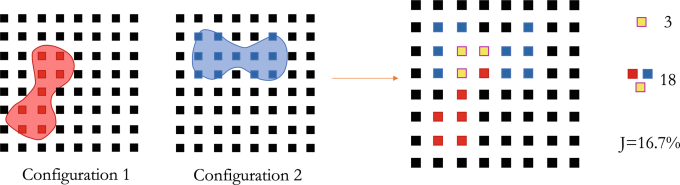
The next, and novel, step was applying the Jaccard index to verify that the configurations were distinct. The Jaccard index measures similarity between sample sets, and the team used it to quantify spatial similarity between cell configurations by mapping the biconcave RBC shapes to grid points, then calculating the configurations’ shared points. (See Figure 2.) This process resulted in numerical characterization of a set of cell configurations.
Past a certain point, increasing the number of configurations did not significantly change their probability distribution. In fact, Draeger notes, “We saw the distribution become less noisy with higher numbers, which gave us confidence that, as a first pass, a sufficient ensemble size for our parameters is 72 configurations.”
Randles adds, “Due to the stochastic nature of RBC distribution, sampling the full ensemble of potential distributions to assess cellular behavior is important. This paper takes an important step in the process by establishing metrics we can use to compare different starting positions of the cells and drive optimal selection of configurations that must be tested.” As ensemble studies can help reduce simulation costs, smarter sampling of cell configurations—like the approach detailed in the team’s paper—could ultimately lead to more efficient use of expensive simulations and more confidence in their accuracy.
Past, Present, and Future
CASC’s relationship with Duke University originated when Randles developed a blood flow dynamics code called HARVEY during her Lawrence Fellowship (2013–2015), which Draeger helped scale to run on the Lab’s supercomputers. “Historically, medical simulations had not harnessed HPC power very well, so they were often limited by being run on small clusters or desktops,” Draeger notes.
Randles joined Duke in 2015 where she is now the Alfred Winborne and Victoria Stover Mordecai Assistant Professor of Biomedical Sciences, leading the Randles Lab within the Pratt School of Engineering. In the years since, she and Draeger have advised Duke students on several biomedicine-meets-HPC projects.
The collaborators have observed the use of HPC in biomedical research grow over time, such as to gain insights into disease progression or drug response. Roychowdhury points out, “HPC developments can unlock advances in biomedicine, while applications in biomedical computing drive the need for innovative HPC design. It’s fascinating how intertwined these two fields have become.”
Indeed, with advances in fluid structure interaction models, Randles states, “We are now able to use leadership-class systems to simulate hundreds of millions of individual RBCs at a time. Now that these models are attainable, we are faced with the bigger question of how to use them to draw meaningful conclusions about the underlying biological phenomena.”
In CASC, cell interactions inside a blood vessel is just one example of physical phenomena that researchers explore through simulation. Physics simulations of small particles—atoms, cells, and so on—are useful in many domains to represent much larger systems of particles. With the success of the team’s paper, Draeger looks beyond the team’s application to other scenarios that aggregate average behavior with multiple simulations. “As you solve HPC problems in one space, you often come up with techniques, solutions, and ideas that feed back into other spaces,” he explains. “We’re building an ecosystem of HPC expertise and science expertise that’s beneficial to the Lab as a whole as well as to our academic partners.”
Learn more about research using HARVEY and blood flow modeling:
- Using models, 3D printing to study common heart defect
- Research team pairs 3D bioprinting and computer modeling to examine cancer spread in blood vessels
- Lab team develops first-ever living 3D-printed aneurysm to improve surgical procedures, personalize treatments
- 2015 Gordon Bell Prize finalist: Randles named finalist for supercomputing’s top honor
—Holly Auten