Scientists at Lawrence Livermore have grappled with a number of challenges in their attempts to develop sustainable computational workflows: running high performance computing software can be complex and error prone; external solutions take time to vet and usually employ technology that violates Livermore security policies; other “black box” tools can be inflexible and cumbersome, hindering users from developing customized methodologies and hiding implementation.
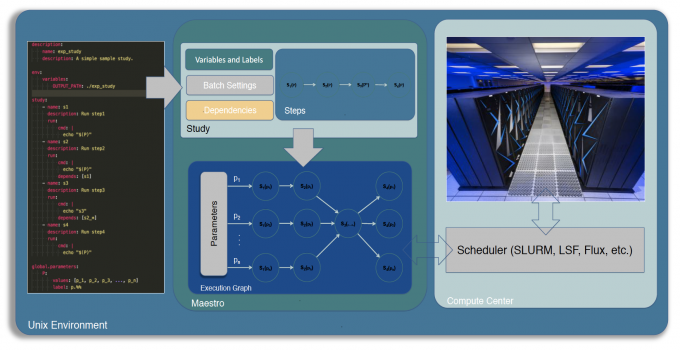
To address these challenges, computer scientist Frank Di Natale led the development of the Maestro Workflow Conductor—a lightweight, open-source Python tool that can launch multi-step software simulation workflows in a clear, concise, consistent, and repeatable manner—locally as well as on supercomputers. Maestro’s development emerged from the need for a general workflow software system that could deliver functionality to an array of disciplines ranging from machine learning to molecular dynamics, cancer, and cardiac research.
Ideally, scientific workflows allow scientists to document and execute a series of multi-step specifications or computational tasks, and determine the dependencies between these tasks when they want to run a simulation. The essential function of a scientific workflow is to manage and organize data in a variety of ways from short serial dependencies to more complex predictive aggregations. Available computational workflow tools, however, don’t allow LLNL scientists to effectively and easily execute their studies. Maestro has changed that.
Di Natale likens the Maestro user experience to a science notebook that documents all the designated variables, steps, and parameters as one would run a physical experiment. Users simply need to mock up their workflow in YAML following an easy-to-use format and Maestro will handle the scheduling, script generation, variable substitution, and then run and manage the study—generating data-rich results that allow users to “focus on the science” rather than get bogged down in coding.
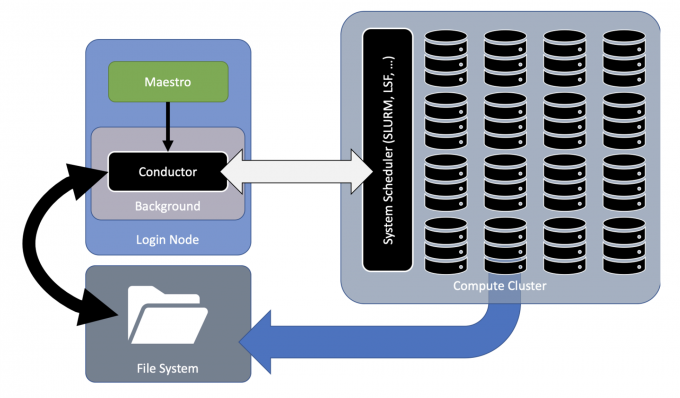
Maestro is unique because it allows users to customize their multi-step workflows as well as their post-processing—enhancing efficiency and productivity. It’s lightweight because it minimizes dependency on external and heavyweight software packages, and is designed to interface with multiple schedulers, so users can run their workflow with the software stack native to the machine running it.
For Di Natale, one of the greatest challenges has been encouraging generality in a community working in many domains and specialties often driven by urgent, mission-critical milestones that don’t necessarily lend themselves to the development of long-term, sustainable workflow infrastructure. “I found that projects were solving the same problems again and again, and that users were applying similar solutions to specific problems but using different platforms,” says Di Natale.
Implementing Maestro has meant teaching users a new, shared vocabulary that allows them to discuss and describe their work with each other. Once users have mastered the vocabulary and translated their workflow to a study specification, they can fine-tune their workflows, add or delete parameters, and repeat complete workflows.
Because Maestro users now communicate, define, and run their workflows in a uniform way, studies can be stored, built upon, shared, and integrated into larger systems. When using Maestro, each study is a template for repeatedly running a set of parameters. “Before Maestro, it took a long time to stand up new workflows. Maestro has changed that by providing a consistent framework that can break down workflows into smaller pieces, and facilitate automated execution,” explains Di Natale.
Many LLNL scientists have adopted Maestro since its inception and are now using it to support L1 (level 1) and L2 (level 2) milestones. And as one of the next-generation tools for LLNL’s global security response efforts, Maestro will facilitate the prompt delivery of mission critical simulations and calculations.
Tom Desautels, a data scientist in LLNL's Computational Engineering’s Machine Learning group, has used Maestro to compute the favorability of antibody/antigen interactions. These calculations support the development and design of antibodies that might combat and neutralize the SARS-CoV-2 virus that causes COVID-19.
Stewart He, a data scientist in LLNL's Global Security-Computing Applications Division (GS-CAD), used Maestro in combination with the ATOM Modeling PipeLine (AMPL) to predict the safety and pharmacokinetic properties of over 26 million drug-like compounds. These predictions will be combined with calculations of the binding affinities between proteins and inhibitors to recommend potential experimental drugs in the battle against COVID-19.
Mikel Landajuela, a postdoctoral researcher in LLNL's Biosciences and Biotechnology Division, has used Maestro to develop a pipeline of cardiac simulations to develop non-invasive imaging of cardiac electrical phenomena using only an electrocardiogram (ECG) as input—which led to a patent. “Maestro is the perfect tool to organize your workflow; it takes care of the tedious part of job scheduling while allowing a high degree of flexibility and user customization,” says Landajuela.
Looking ahead, Di Natale envisions expanding Maestro’s capabilities to what he’s termed “symphonies”—the program’s ability to facilitate a decision-making loop that can link to and work with other studies. “At a higher level, if each study is considered a unit, then we can plan workflows around them. This means that common problems such as optimization, UQ, and machine-learned decision making can influence whether a study needs to be run over more parameters,” says Di Natale.
Di Natale’s goal in developing Maestro “was to introduce a level of grace into how we talk about our work, and how we perform computational science.” He hopes to change the way LLNL and the computational science community thinks about their workflows, enabling open communication and shared collaboration that support scientific advances.
Maestro is open source and available on GitHub as well as on LLNL’s software catalog. It’s easily installed, and users should be up in running in a matter of minutes.
Maestro’s development is primarily funded by the Pilot 2 Cancer Moonshot in conjunction with RADIUSS. Maestro was initially funded by the Exascale Computing Project Workflow Project and the Institutional Center of Excellence.