Disclaimer: This article is more than two years old. Developments in science and computing happen quickly, and more up-to-date resources on this topic may be available.
The IEEE international eScience conference, which emphasizes compute- and data-intensive research methods, bestowed the 2022 Best Paper Award on a multidisciplinary team that includes LLNL staff. The paper, “Scalable Composition and Analysis Techniques for Massive Scientific Workflows,” details the optimization of a drug screening workflow for the American Heart Association (AHA). The AHA Molecule Screening (MoleS) workflow combines specialized software tools to manage high performance computing (HPC) hardware heterogeneity.
The HPC landscape is shifting toward more heterogeneity with different parts of a scientific workflow running on specialized computing resources. Such integrations are becoming more complex and difficult to manage. “Massive scientific workflows require HPC management solutions that don’t currently exist,” notes computer scientist Daniel Milroy. “The paper describes the complexity of integrating interdisciplinary science with different computing fields, including the convergence of traditional and cloud computing, and acknowledges what will be required to conduct large-scale science in the future.”
Billions of Calculations
The AHA engaged LLNL to develop a queryable atlas of human proteins and small-molecule compounds, aimed at better understanding drug side effects. “A drug compound binds with an average of 32 protein targets when metabolizing throughout the human body, sometimes causing severe side effects,” explains computational chemist Xiaohua Zhang. “The atlas will help researchers study on- and off-target binding of each compound in order to better understand these side effects.”
The atlas is a massive database containing more than 28,000 structural models for nearly 15,000 human proteins, as well as 2.1 million small-molecule compounds. Calculating the billions of interactions between all of the proteins and compounds is a huge, complicated computational undertaking. Zhang’s team in LLNL’s Biosciences and Biotechnology Division involved Computing’s HPC experts from the project’s beginning. He states, “We knew we needed a workflow manager to handle all the calculations.”
Composite, Therefore Complex
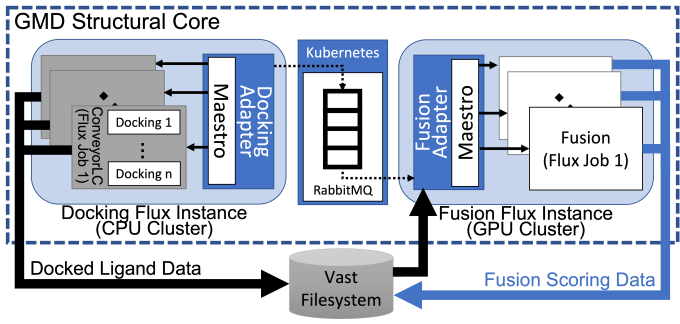
The MoleS end-to-end workflow relies on both general-purpose and domain-specific software tools, some of which are open source and/or developed at LLNL: Maestro for workflow execution, Flux for workload management and scheduling, RabbitMQ for message brokering (orchestrated by Kubernetes), ConveyorLC for docking automation tasks, Fusion machine learning algorithms for binding affinity predictions, and GMD (Generative Molecular Design) for the small-molecule discovery loop.
In a composite workflow like MoleS, tasks are generated, scheduled, and completed by these software components. But each component has different computational requirements, which makes measuring and optimizing the full workflow’s performance challenging. The MoleS workflow uses LLNL’s CPU-based Ruby supercomputer for molecule batch creation and completion as well as ConveyorLC and Docking jobs. The CPU-GPU Lassen supercomputer handles Fusion machine learning jobs—which consume Docking data—while the RabbitMQ server runs on Livermore Computing’s on-premises cloud persistent data services cluster running Red Hat OpenShift. “Ruby runs the more traditional HPC application, and Lassen runs the machine learning analysis. The Kubernetes cluster facilitates communications between the two,” Milroy explains. The workflow must be smart enough to coordinate the handoff points and broker task messages.
Combining all of these components, the team created composition and analysis techniques that optimize and scale scientific workflows for HPC centers. This approach defines the scope of variables that impact the workflow’s performance, so researchers can determine opportunities for improvements and efficiencies. As demonstrated by the MoleS project—where performance increased by a factor of up to 2.45x—the techniques significantly enhanced the team’s ability to create a high performance composite workflow.
Managing the Critical Path
A scientific workflow must complete tasks in the proper order as quickly as possible, so minimizing the tasks’ critical paths becomes essential. Moreover, the choice of performance variables and their individual optimization needs can affect overall workflow performance. Additionally, automation of cross-system tasks can help streamline trial-and-error during performance tuning.
The paper outlines the steps for identifying performance variables and their dependent relationships, and proposes a consistent way to gather performance data that includes the critical path. Furthermore, the team leverages an LLNL-developed open-source tool analysis tool called PerfFlowAspect that allows researchers to monitor various parts of a workflow. PerfFlowAspect captures performance characteristics of the workflow during runtime and writes the annotated data to a standard file format for consumption by a visualization tool such as Perfetto. Critical-path events can change as the workflow progresses, and the team’s strategy makes possible continuous optimization.
Team Science
The team’s accomplishment provides a springboard for new applications and techniques, as the MoleS workflow paradigm is generalizable to other domains. For instance, Zhang also contributes to a multiscale simulation project that leverages a similar workflow architecture. Additionally, the MoleS team is exploring workflow migration to an Amazon Web Services cloud platform using Docker containers, and a project funded under the Laboratory Directed Research and Development Program is developing dynamic computing resource allocation based on machine learning models.
“Our multidisciplinary collaboration has been very productive,” Zhang points out. “Individually, our work requires expertise outside our own domains. Together we can create a meaningful workflow with good results that can be adapted by other researchers and used in future projects.”
Along with Zhang and Milroy, the paper’s co-authors are LLNL researchers Dan Kirshner, Bronis de Supinski, Brian Van Essen, Jonathan Allen, and Felice Lightstone; former LLNL staff Dong Ahn (lead author), Stephen Herbein, Francesco Di Natale, Ian Karlin, and Sam Ade Jacobs; and Jeffrey Mast from Teres Technologies.
—Holly Auten