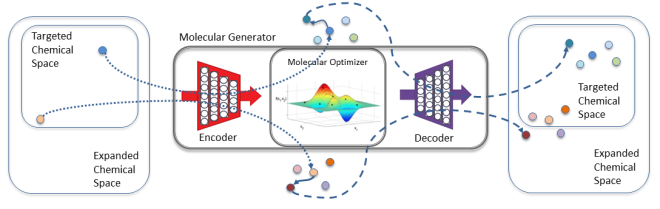
The engine behind the COVID-19 antiviral drug design pipeline is the GMD software, which combines a computational autoencoder framework with sophisticated ML algorithms to propose molecular structures, identify those with desirable properties, and suggest new molecules based on the best results.
The pipeline produces incremental improvements through GMD, which can be conceptualized as a loop: A library of molecules is scored against design criteria, the best-scoring molecules are combined and altered to create new molecules, then the new molecules are evaluated.
“The loop is repeated for multiple generations, evolving the population of molecules toward a set with the desired properties,” McLoughlin explains. “With this technology, we iteratively improve a set of molecular designs.”
The team’s autoencoder solution is an ML algorithm that compresses data—in this case, molecular structures—for analysis as vectors in the latent space. Not to be confused with viral vectors, these vectors are mathematical representations of molecules. In other words, McLoughlin says, “Our models translate a chemical structure into a set of numbers.”
A latent vector can be manipulated to create a new molecule. Alternatively, McLoughlin states, “Pairs of high-scoring latent vectors can have their elements shuffled together to form a new vector, much as genes from a father and mother are recombined to create a child’s genome.” Over hundreds of loop generations, molecules with the best chance of experimental success emerge.
The final piece of this complex computational puzzle is the LBANN software, which is optimized for huge datasets and scales ML models to train on large HPC systems. The team used LBANN to train an autoencoder on the dataset of 1.6 billion molecules—nearly an order of magnitude more molecules than in any other work reported to date. Running on 4,160 nodes (each with 4 GPUs) of the 125-petaflop Sierra supercomputer with 97.7% efficiency, the model’s training time fell from a day to just 23 minutes.
The achievement combines three unique factors: an unprecedented dataset, a powerful HPC system, and complex deep-learning software. Jacobs states, “No one has trained a deep-learning model on this amount of molecular data before. This work is the first demonstration of LBANN at a billion-plus scale, as well as the first time scaling to all of Sierra’s nodes.”
The autoencoder training method was named a finalist for the 2020 Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research, which recognizes computational performance and innovation alongside contributions toward understanding the disease.