LLNL is delivering mission impact with AI expertise, including for biodefense and national security materials. As bio threats emerge, traditional methods of fighting them are no longer sufficient and new approaches are needed for rapid antibody development. Computational prediction of protein structure enables many applications in this mission space.
The Need for Open Weight Inference Models
Deep learning has revolutionized protein folding with commercial AI models such as AlphaFold. However, AlphaFold models prohibit many use cases, including distillation of datasets or derivative models. Experimental datasets, while providing ground truth, are not sufficient for training modern models, which require massive number of high quality training samples that are now only available through cross- and self-distillation approaches.
For national security, it is critical for LLNL to be able to evolve state-of-the-art models to meet future needs. Distillation—using a pretrained model to produce high-quality data for further training—and its resulting datasets are critical for future-proofing protein folding models. Moreover, open models, like OpenFold from Columbia University, spur innovation and enable new insights.
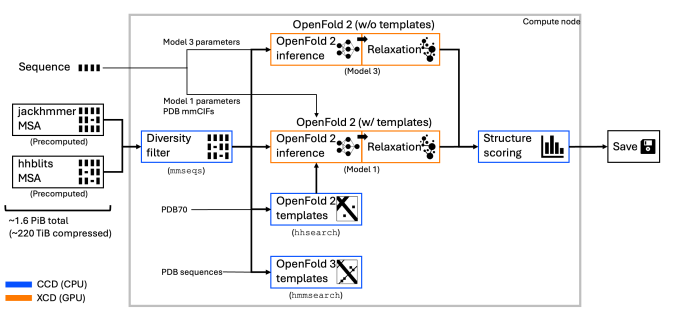
ElMerFold, a high performance framework for large-scale inference and distillation on Livermore supercomputers with OpenFold-specific optimizations, is LLNL’s answer to these mission needs. It builds on our combined leadership in HPC and AI and our long tradition of successful open-source work.
OpenFold Meets the El Capitan Generation of Machines
ElMerFold is a high performance framework for large-scale inference and distillation on El Capitan with OpenFold-specific optimizations. The ElMerFold team is composed of staff from Livermore Computing and the Center for Applied Scientific Computing as well as AMD engineers to execute:
- AMD Instinct™ MI300A Acceleratoroptimizations
- LBANNv2: memory optimizations
- Efficient kernel implementations
- On-node optimizations
- Workflow orchestration
- Persistent inference servers
- I/O optimizations
- I/O offload to Rabbits
- Streaming data (de)compression
- Startup optimizations
These tunings provided an improved environment for the OpenFold3 monomer distillation workflow.
For more details on the technical aspects of ElMerFold, see ElMerFold: Exascale Distillation Workflows for Protein Structure Prediction on El Capitan.
Scaling Up: Improving the Model Improves the Machine
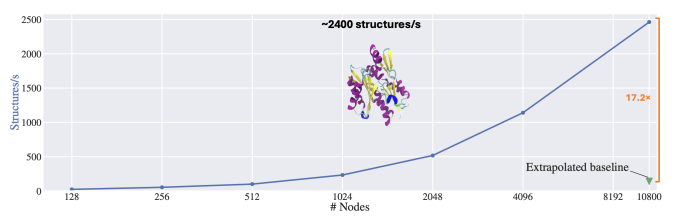
ElMerFold is a prime example of HPC-powered data generation at scale. Leveraging LLNL’s El Capitan and Tuolumne supercomputers, scientists generated over 41 million protein structure predictions for OpenFold3’s self-distillation training. The framework delivered structure predictions on the AMD APUs at the heart of our newest systems 17x faster than the original inference pipeline, making massive dataset builds plausible.
However, these models were distilled during El Capitan’s open-science “shakeout” phase and improved not only the models but also the machine. For example, ElMerFold workloads served as a comprehensive stress test for the machine, helping uncover hardware bottlenecks, validate end-to-end APU and interconnect performance, and refine system configurations. This real-world debugging accelerated readiness for production science runs and informed both LLNL and AMD on where to tune El Capitan for peak reliability.
Additionally, pushing millions of APU/GPU-accelerated inferences on El Capitan drove fixes that benefit all NNSA workloads—both AI and more traditional modeling and simulation. The project was an early large-scale user of the HPE Rabbit configurable near-node storage capability on El Capitan, also driving fixes and improvements to the benefit of all future work. Finally, the LLNL/AMD collaboration yielded optimizations that are now being applied to the software stack, including memory management and data movement.